
SysFate
Systems Biology of cell Fate decisions

Marco Antonio Mendoza
Laboratory of Systems Biology
ISSB laboratory; UMR 8030
Génomique Métabolique

Our laboratory study cell fate decisions from a systems biology perspective, i.e. by aiming to understand their transitions from the reorganization of complex regulatory wires defining their biological state.

Our ambition is focused on enhancing our understanding of cell fate transitions in a variety of biological systems by the reconstruction of the gene regulatory networks that are reorganized during these transition events. At term we aim to provide a comprehensive view of the gene regulatory wires defining each of the cell/tissue types in an organism, thus representing a landmark catalog for the future therapeutic developments in regenerative medicine.
In this context, we follow the innovation engine strategy of Systems biology; i.e. We address biological questions with the help of novel technological developments and bioinformatics.
From big-data functional genomics to the reconstruction of regulatory programs defining biological systems
With the democratization of sequencing technologies, the major challenge of the present genomics era does not reside anymore in data acquisition, but rather in having computational solutions for interrogating billions of data points from several datasets in a comparative manner such that their embedded information might provide means to describe biological systems.
Over the years, our team addressed these questions through the development of a universal quality control strategy for ChIP-seq and related datasets (Mendoza-Parra et al; NAR 2013), which has been applied to certify > 100,000 public datasets and more recently by the release of a user-friendly online resource for ultrafast retrieval, visualization, and comparative analysis of tens of thousands of genomics datasets to gain new functional insight from global or focused multidimensional data integration.
The innovation engine strategy in Systems biology
Main research projects:

n° Citations: 2975; h-index: 21; i10-index: 29 (December 2025)
2025
-
A. Trajkova, E. Zakiev, M. Mendoza-Ferri, I. Messahli, M. Ruel, I. Krossa, Y. Tapponnier, E. Dejonckere, E. Hunter, L. Claret, B. Martin, N. Gadot, F. Forest, C. Heinrich, N. Martin, D. Bernard, Y. Grinberg-Bleyer, M. Mendoza-Parra, A. Huyghe, F. Lavial. Pioneer Transcription Factors Differentially Co-opt Bcl11a and Bcl11b to orchestrate Reprogramming; BioRxiv December 22, 2025.
-
Kowar A, Becker JP, Del Pizzo R, Tang Z, Champagne J, Wellach K, Samimi K, Galindo-Albarrán A, Körner PR, Montenegro Navarro J, Elía A, Tilghman FM, Sakeer H, Mendoza-Parra MA, Riemer AB, Agami R, Loayza-Puch F. Upstream open reading frame translation enhances immunogenic peptide presentation in mitotically arrested cancer cells. Nat Commun. 2025 Aug 27;16(1):8008. doi: 10.1038/s41467-025-63405-2. PMID: 40866359; PMCID: PMC12391379.
-
Mendoza-Ferri MG, Lozachmeur G, Duvina M, Perret L, Merciris D, Gigout A, Mendoza-Parra MA. Tissular chromatin-state cartography based on double-barcoded DNA arrays that capture unloaded PA-Tn5 transposase. Genome Res. 2025 Jun 6. doi: 10.1101/gr.280305.124. Epub ahead of print. PMID: 40360185.
-
Ariel Galindo-Albarrán, Aysis Koshy, Maria Grazia Mendoza-Ferri, Marco Antonio Mendoza-Parra, Decoding transcriptional identity during neuron-astroglia cell fate driven by RAR-specific agonists, NAR Molecular Medicine, Volume 2, Issue 2, April 2025, ugaf018
-
Megat S, Marques C, Hernán-Godoy M, Sellier C, Stuart-Lopez G, Dirrig-Grosch S, Gorin C, Brunet A, Fischer M, Keime C, Kessler P, Mendoza-Parra MA, Zwamborn RAJ, Veldink JH, Scholz SW, Ferrucci L, Ludolph A, Traynor B, Chio A, Dupuis L, Rouaux C. CREB3 gain of function variants protect against ALS. Nat Commun. 2025 Mar 26;16(1):2942. doi: 10.1038/s41467-025-58098-6. PMID: 40140376; PMCID: PMC11947196.
-
Ariel Galindo-Albarran, Maximilien Duvina, Marco Antonio Mendoza-Parra; Enhancing spatial omics resolution by pseudo-interstitial pixels inference; January 27, 2025; BioRxiv
2024
-
Galindo-Albarran, Aysis Koshy, Maria Grazia Mendoza-Ferri, Marco A Mendoza-Parra; Decoding transcriptional identity during Neuron-Astroglia Cell Fate driven by RAR-specific agonists; December 24th 2024; BioRxiv.
-
Salim Megat, Christine Marques, Marina Hernan Godoy, Chantal Sellier, Geoffrey Stuart-Lopez, Sylvie Dirrig-Grosch, Charlotte Gorin, Aurore Brunet, Mathieu Fischer, Celine Keime, Pascal Kessler, Marco Antonio Mendoza-Parra, Sonja W Scholz, Luigi Ferrucci, Albert Ludolph, Bryan Traynor, Adriano Chio, Luc Dupuis, Caroline Rouaux; CREB3 gain of function variants protect against ALS; October 11th 2024; BioRxiv
-
Maria B. Goncalves, Yue Wu, Earl Clarke, John Grist, Julien Moehlin, Marco Antonio Mendoza-Parra, Carl Hobbs, Barret Kalindjian, Henry Fok, Adrian P. Mander, Hana Hassanin, Daryl Bendel, J¨org T¨aubel, Tim Mant, Thomas Carlstedt, Julian Jack and Jonathan P. T. Corcoran. C286, an orally available retinoic acid receptor β agonist drug, regulates multiple pathways to achieve spinal cord injury repair. Frontiers Molecular NeuroScience. 2024; 20th August 2024; doi: 10.3389/fnmol.2024.1411384
2023
-
Koshy AM, Mendoza-Parra MA. Retinoids: Mechanisms of Action in Neuronal Cell Fate Acquisition. Life. 2023; 13(12):2279. https://doi.org/10.3390/life13122279
-
Antoine Aubert, Maria Grazia Mendoza-Ferri, Aude Bramoulle, François Stüder, Bruno Maria Colombo, Marco Antonio Mendoza-Parra; PSEN1-M146V and PSEN1-A246E mutations associated with Alzheimer’s disease impair proper microglia differentiation; October 10, 2023; BioRxiv.
-
Gwendoline Lozachmeur, Aude Bramoulle, Antoine Aubert, François Stüder, Julien Moehlin, Lucie Madrange, Frank Yates, Jean-Philippe Deslys, Marco Antonio Mendoza-Parra; Three-dimensional molecular cartography of human cerebral organoids revealed by double-barcoded spatial transcriptomics; Cell Reports Methods; 1st Sept. 2023.
-
Xian-Yang Qin, Marco Antonio Mendoza-Parra and Yohei Shirakami; Editorial: Cancer and nutrients: new chemicals, signals, and biomarker-based therapy; Frontiers in Oncology; April 11, 2023.
2022
-
Aysis Koshy, Elodie Mathieux, François Stüder, Aude Bramoulle, Michele Lieb, Bruno Maria Colombo, Hinrich Gronemeyer, Marco Antonio Mendoza-Parra; Synergistic activation of RARb and RARg nuclear receptors restores cell specialization during stem cell differentiation by hijacking RARa-controlled programs; Life Science Alliance; November 29th 2022
-
A. Huyghe, G. Furlan, J. Schroeder, E. Cascales, A. Trajkova, M. Ruel, F. Stüder, M. Larcombe, Y. Bo Yang Sun, F. Mugnier, L. De Matteo, A. Baygin, J. Wang, Y Yu, N. Rama, B. Gibert, J. Kielbassa, L. Tonon, P. Wajda, N. Gadot, M. Brevet, M. Siouda, P. Mulligan, R. Dante, P. Liu, H. Gronemeyer, M. Mendoza-Parra, J.M. Polo and F. Lavial; Comparative roadmaps of reprogramming and oncogenic transformation identify Bcl11b and Atoh8 as broad regulators of cellular plasticity; Nature Cell Biology; September 8th 2022.
2021
-
Akinchan Kumar, Yasenya Kasikci, Alaa Badredine, Karim Azzag, Marie L Quintyn Ranty, Falek Zaidi, Nathalie Aragou, Catherine Mazerolles, Bernard Malavaud, Marco A Mendoza-Parra, Laurence Vandel, Hinrich Gronemeyer; Patient-matched analysis identifies deregulated networks in prostate cancer to guide personalized therapeutic intervention; Am J Cancer Res; 2021 Nov 15;11(11):5299-5318. eCollection 2021
-
Aubert, Antoine, François Stüder, Bruno M. Colombo, and Marco A. Mendoza-Parra ; A Core Transcription Regulatory Circuitry Defining Microglia Cell Identity Inferred from the Reanalysis of Multiple Human Microglia Differentiation Protocols; Brain Sciences 11, no. 10: 1338 ; 2021.
-
Julien Moehlin, Aysis Koshy, François Stüder and Marco Antonio Mendoza-Parra; Protocol for using MULTILAYER to reveal molecular tissue substructures from digitized spatial transcriptomes; STAR Protocols; September 17, 2021.
-
François Stüder, Jean-Louis Petit, Stefan Engelen, Marco Antonio Mendoza-Parra; Real-time SARS-CoV-2 diagnostic and variants tracking over multiple candidates using nanopore DNA sequencing; Scientific Reports; 11; 15869; 5 August 2021.
-
Julien Moehlin, Bastien Mollet, Bruno Maria Colombo, Marco A. Mendoza-Parra, Inferring biologically relevant molecular tissue substructures by agglomerative clustering of digitized spatial transcriptomes with multilayer; Cell Systems; 07 May 2021.
-
Florent Colin, Kristine Schauer, Ali Hamiche, Pierre Martineau, Jean‐Paul Borg, Jan Bednar, Guilia Bertolin, Luc Camoin, Yves Collette, Stephan Dimitrov, Isabelle Fournier, Vincent Hyenne , Marco A. Mendoza‐Parra, Xavier Morelli, Philippe Rondé, Izabela Sumara, Marc Tramier, Patrick Schultz, Jacky G. Goetz, The NANOTUMOR consortium ‐ towards the Tumor Cell Atlas; Biology of the Cell; 07 February 2021.
2020
-
Maha Siouda, Audrey D. Dujardin, Laetitia Barbollat-Boutrand, Marco A. Mendoza-Parra, Benjamin Gibert, Maria Ouzounova, Jebrane Bouaoud, Laurie Tonon, Marie Robert, Jean-Philippe Foy, Vincent Lavergne, Serge N. Manie, Alain Viari, Alain Puisieux, Gabriel Ichim, Hinrich Gronemeyer, Pierre Saintigny, Peter Mulligan. CDYL2 epigenetically regulates MIR124 to control NF-κB/STAT3-dependent breast cancer cell plasticity, iScience. 2020 May 6.
2019
-
Blum M, Cholley PE, Malysheva V, Nicaise S, Moehlin J, Gronemeyer H, Mendoza-Parra MA. A comprehensive resource for retrieving, visualizing, and integrating functional genomics data. Life Sci Alliance. 2019 Dec 9;3(1).
-
Goncalves, M.B., Moehlin, J., Clarke, E., Grist, J., Hobbs, C., Carr, A.M., Jack,J., Mendoza-Parra, M.A., Corcoran, J.P.T, RARβ agonist drug (C286) demonstrates efficacy in a pre-clinical neuropathic pain model restoring multiple pathways via DNA repair mechanisms, iScience. 2019 Nov 22;21:562-563.
-
Elodie Mathieux and Marco Antonio Mendoza-Parra; Systems Biology Perspectives for Studying Neurodevelopmental Events [Online First], IntechOpen; April 26th 2019; DOI: 10.5772/intechopen.85072. Available from: https://www.intechopen.com/online-first/systems-biology-perspectives-for-studying-neurodevelopmental-events
2018
-
Lucas Fauquier, Karim Azzag, Marco Antonio Mendoza Parra, Aurélie Quillien, Manon Boulet, Sarah Diouf, Gilles Carnac, Lucas Waltzer, Hinrich Gronemeyer & Laurence Vandel; CBP and P300 regulate distinct gene networks required for human primary myoblast differentiation and muscle integrity; Scientific Reports; 2018 August 22.
-
Pierre-Etienne Cholley, Julien Moehlin, Alexia Rohmer, Vincent Zilliox, Samuel Nicaise, Hinrich Gronemeyer and Marco Antonio Mendoza-Parra; Modeling gene-regulatory networks to describe cell fate transitions and predict master regulators; Nature Systems Biology & Applications; 2018 August 2.
-
Shinya Fujii, Shuichi Mori, Hiroyuki Kagechika, Marco Antonio Mendoza Parra and Hinrich Gronemeyer; Development of biotin-retinoid conjugates as chemical probes for analysis of retinoid function; Bioorganic & Medicinal Chemistry Letters; 2018 June 6,
-
Maja Milanovic, Dorothy N.Y. Fan, Dimitri Belenki, J. Henry M. Däbritz, Zhen Zhao, Yong Yu, Jan R. Dörr, Lora Dimitrova, Dido Lenze, Ines A. Monteiro Barbosa, Marco A. Mendoza-Parra, Tamara Kanashova, Marlen Metzner, Katharina Pardon, Maurice Reimann, Andreas Trumpp, Bernd Dörken, Johannes Zuber, Hinrich Gronemeyer, Michael Hummel, Gunnar Dittmar, Soyoung Lee and Clemens A. Schmitt; Senescence-associated reprogramming promotes cancer stemness; Nature; 2018 Jan 4;553 (7686):96-100.
2017
-
MAM Saleem, Marco Antonio Mendoza-Parra, PE Cholley, M Blum and Hinrich Gronemeyer; Epimetheus: a multi-profile normalizer for epigenomic sequencing data; BMC Bioinformatics; 12 May 2017, 18 (1), 259;. [IF 2.435]
2016
-
Marco Antonio Mendoza-Parra, Valeriya Malysheva, Mohamed Ashick Mohamed Saleem, Michele Lieb, Aurelie Godel and Hinrich Gronemeyer; Reconstructed cell fate-regulatory programs in stem cells reveal hierarchies and key factors of neurogenesis; Genome Research; 20 September 2016, 26 (11), 1505-1519. [IF 14.630]
-
Valeriya Malysheva, Marco Antonio Mendoza-Parra, Mohamed Ashick Mohamed Saleem and Hinrich Gronemeyer; Reconstructing gene regulatory networks of tumorigenesis; Genome Medicine; 19 May 2016, 8 (1), 57. [IF 5.338]
-
Marco Antonio Mendoza-Parra, Matthias Blum, Valeriya Malysheva, Pierre-Etienne Cholley and Hinrich Gronemeyer; LOGIQA: A database dedicated to Long-range Genome Interactions Quality Assessment; BMC Genomics; 16 May 2016, 17 (1), 355. [IF 3.986]
-
Marco A. Mendoza-Parra, Mohamed Saleem MA, Matthias Blum, Pierre-Etienne Cholley and Hinrich Gronemeyer; NGS-QC Generator: A Quality control system for ChIP-seq and related deep sequencing-generated datasets; Statistical Genomics : Methods and Protocols; Springer Book Series; 24 March 2016; Volume 1418; pp 243-65.
-
Marco Antonio Mendoza-Parra, Vincent Saravaki, Pierre-Etienne Cholley, Matthias Blum, Benjamin Billoré and Hinrich Gronemeyer; Antibody performance in ChIP-sequencing assays: From quality scores of public data sets to quantitative certification; F1000Research; 12 January 2016; 5:14; doi: 10.12688/f1000research.7637.2
2015
-
Marco Antonio Mendoza-Parra, and Hinrich Gronemeyer, Systems biology approaches to uncover the function of RAR and RXR subtypes and of selective ligands in gene regulation during cell lineage specification, The Retinoids: Biology, Biochemistry and Disease" (Wiley-Blackwell publisher Book Series); May 2015. pp 165-192.
-
Ronan Chaligné, Tatiana Popova, Marco Antonio Mendoza Parra, Mohamed Ashick Mohamed Saleem, David Gentien, Kristen Ban, Tristan Piolot, Olivier Leroy, Odette Mariani, Hinrich Gronemeyer, Anne Vincent-Salomon, Marc-Henri Stern and Edith Heard ; The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer; Genome Research; 2015 Apr, 25(4): 488-503. [IF 14.630]
2014
-
Marco A. Mendoza-Parra and Hinrich Gronemeyer; Assessing quality standards for ChIP-seq and related massive parallel sequencing-generated datasets: When rating goes beyond avoiding the crisis; Genomics Data; Dec. 2014; vol. 2, pages 268-273.
-
Jelena Vjetrovic, Pattabhiraman Shankaranarayanan, Marco A. Mendoza-Parra and Hinrich Gronemeyer; Senescence-secreted factors activate Myc and sensitize pretrasformed cells to TRAIL-induced apoptosis; Aging Cell; 2014 June; 13(3):487-96; doi: 10.1111/acel.12197 [IF. 6.340]
-
Marco Antonio Mendoza-Parra, and Hinrich Gronemeyer, Integrative genomics to dissect retinoids functions, The Biochemistry of Retinoic Acid Receptors I: Structure, Activation and Function at the Molecular Level; Subceullar Biochemistry; Springer Book Series; 25th June 2014; Volume 70, pp 181-202.
2013
-
Marco Antonio Mendoza-Parra, Wouter Van Gool, Mohamed Saleem MA, Danilo Guillermo Ceschin and Hinrich Gronemeyer; A quality control system for profiles obtained by massive parallel sequencing; Nucleic Acid Research; 2013 Nov 1; 41(21):e196. doi: 10.1093/nar/gkt829. [IF 9.112] (PATENTED)
-
Marco Antonio Mendoza-Parra, Malgorzata Nowicka, Wouter Van Gool and Hinrich Gronemeyer; Characterising ChIP-seq binding patterns by model-based peak shape deconvolution; BMC Genomics; 2013 Nov.26; 14(1): 834. doi: 10.1186/1471-2164-14-834 [IF 3.986]
-
Marco Antonio Mendoza-Parra, and Hinrich Gronemeyer, Genome-wide studies of nuclear receptors in cell fate decisions, Review journal “Seminars in Cell and Developmental Biology”, Thematic issue: Nuclear receptors and cell differentiation, pluripotency, development and stem cells; 2013 Dec; 24 (10-12): 706-15. [IF 6.265]
2012
-
Pattabhiraman Shankaranarayanan*, Marco Antonio Mendoza-Parra*, Wouter Van Gool, Luisa M. Trindade and Hinrich Gronemeyer, Single-tube linear DNA amplification (LinDA) for genome-wide studies using a few thousand cells, Nature Protocols. 2012 Jan 26;7(2):328-38. doi: 10.1038/nprot.2011.447. [IF 9.673] (*equal first author’s contribution)
-
Marco Antonio Mendoza-Parra, Martial Sankar, Mannu Walia and Hinrich Gronemeyer ; POLYPHEMUS : R package for comparative analysis of RNA Polymerase II ChIP-seq profiles by non-linear normalization ; Nucleic Acid Research. 2012 Feb; 40(4):e30 doi: 10.1093/nar/gkr1205. [IF 9.112]
2011
-
Marco Antonio Mendoza-Parra, Mannu Walia, Martial Sankar and Hinrich Gronemeyer ; Dissecting the retinoic acid-induced differentiation of F9 mouse embryonal stem cells through integrative genomic analysis; Molecular Systems Biology; 2011 Oct 11; 7: 538 [IF 10.872]
-
Marco Antonio Mendoza-Parra, Martial Sankar, Mannu Walia and Hinrich Gronemeyer ; POLYPHEMUS : R package for comparative analysis of RNA Polymerase II ChIP-seq profiles by non-linear normalization ; Nucleic Acid Research. 2011 Dec 7; doi: 10.1093/nar/gkr1205. [IF 9.112]
-
Pattabhiraman Shankaranarayanan, Marco Antonio Mendoza-Parra, Mannu Walia, Li Wang, Ning Li, Luisa M. Trindade and Hinrich Gronemeyer, Single-tube linear DNA amplification (LinDA) for robust ChIP-seq, Nature Methods. 2011 Jun 5; 8(7):565-7. [IF 32.072]
-
Silvia Panizza, Marco Antonio Mendoza, Marc Berlinger,Lingzhi Huang, Alain Nicolas, Katsuhiko Shirahige and Franz Klein, Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination, Cell. 2011 Aug 5; 146 (3): 372-83. [IF 32.242]
2009
-
Marco Antonio Mendoza, Silvia Panizza and Franz Klein, Analysis of protein-DNA interactions during meiosis by quantitative chromatin-immunoprecipitation (qChIP), Methods in Molecular Biology Book Series, 2009 ; 557 : 267-83.
non-peer-reviewed articles
-
Marco Antonio Mendoza-Parra, Pattabhiraman Shankaranarayanan & Hinrich Gronemeyer. Sequential chromatin immunoprecipitation protocol for global analysis through massive parallel sequencing (reChIP-seq). Protocol Exchange, DOI 10.1038/protex.2011.1256 (2012).
-
Marco Antonio Mendoza, 2001, Les Phytases : Structure, Caractérisation et Applications, www.123bio.net/revues/
Publications
Networks

RXRa-RARg regulome
Mendoza-Parra et al; Mol. Sys. Biol. 2011
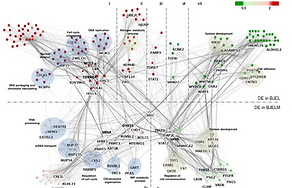
BJ fibroblast stepwise tumorigenesis transformation
Malysheva et al; Genome Medicine. 2016
Master regulatory networks


MEFs to iPS cells reprogramming
TETRAMER
tumorigenic transformation driven by conditioned medium from senescent cells
TETRAMER

retinoids action on neuronal/endodermal cell fate acquisiton
Mendoza-Parra et al; Genome Research. 2016




























